Jelenlegi hely
Quantitative analysis of tubular tree structures

Description
Tubular structures are frequently found in living organisms. The tubes — e.g., arteries or veins — are organized into more complex structures. Trees consisting of tubular segments form the arterial and venous systems, intrathoracic airways form bronchial trees, and other examples can be found.
Computed tomography (CT) or magnetic resonance (MR) imaging provides volumetric image data allowing identification of such tree structures. Frequently, the trees represented as contiguous sets of voxels must be quantitatively analyzed. The analysis may be substantially simplified if the voxel-level tree is represented in a formal tree structure consisting of a set of nodes and connecting arcs. To build such formal trees, the voxel-level tree object must be transformed into a set of interconnected single-voxel centerlines representing individual tree branches. Therefore, the aim of our work was to develop a robust method for identification of centerlines and bifurcation (trifurcation, etc.) points in segmented tubular tree structures acquired in vivo from humans and animals using volumetric CT or MR scanning, rotational angiography, or other volumetric imaging means.
Our method allows to quantitatively analyze tubular tree structures. Assuming that an imperfectly segmented tree was obtained from volumetric data in the previous stages, the presented technique allows to obtain a single-voxel skeleton of the tree while overcoming many segmentation imperfections, yields formal tree representation, and performs quantitative analysis of individual tree segments on a tree-branch basis. The input of the proposed method is a 3D binary image representing a segmented voxel-level tree object. All main components of our method were specifically developed to deal with imaging artifacts typically present in volumetric medical image data. As such, the method consists of the following main steps: (1) airway segmentation, (2) correction of the segmented tree, (3) identification of the tree root, (4) extraction of the 3D centerline—skeletonization, (5) tree pruning, (6) centerline smoothing, (7) identification of branch-points, (8) generation of a formal tree structure, (9) tree partitioning, and (10) quantitative analysis.
The key steps are now illustrated:

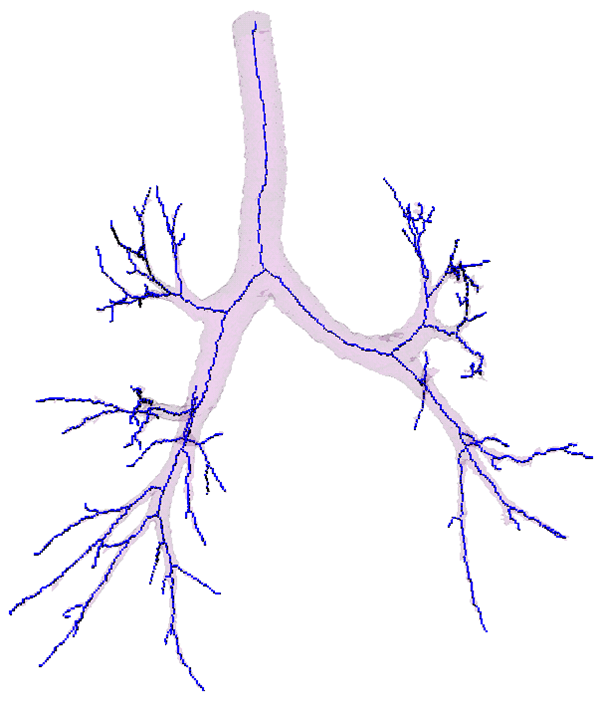
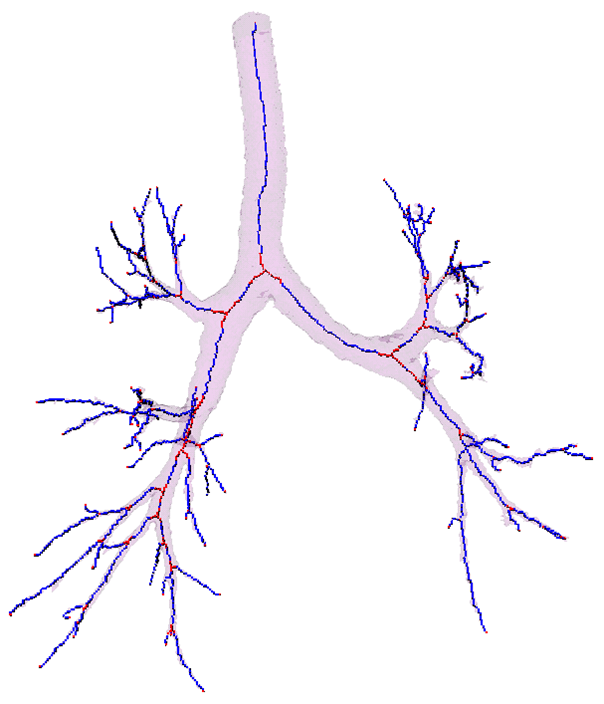
Segmentation - adaptive region growing (left), extracting centerlines - topologically and geometrically correct thinning (middle), and excluding branch-areas based on 3D distance map calculation (right).
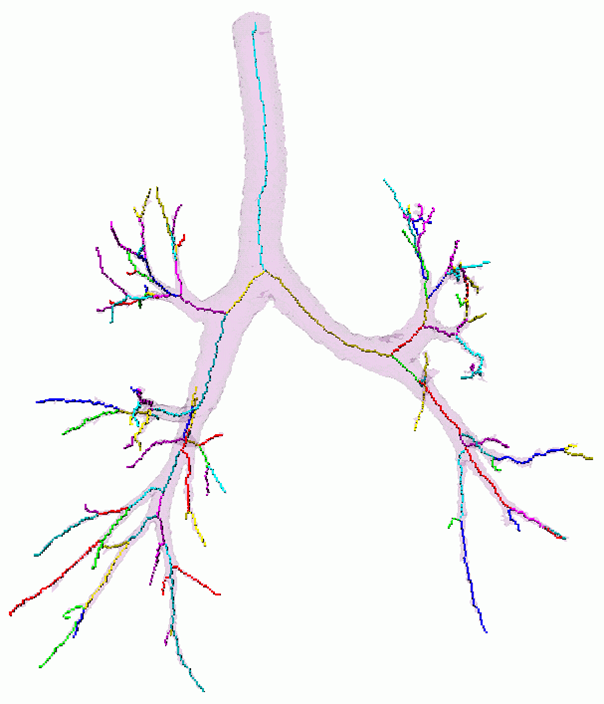
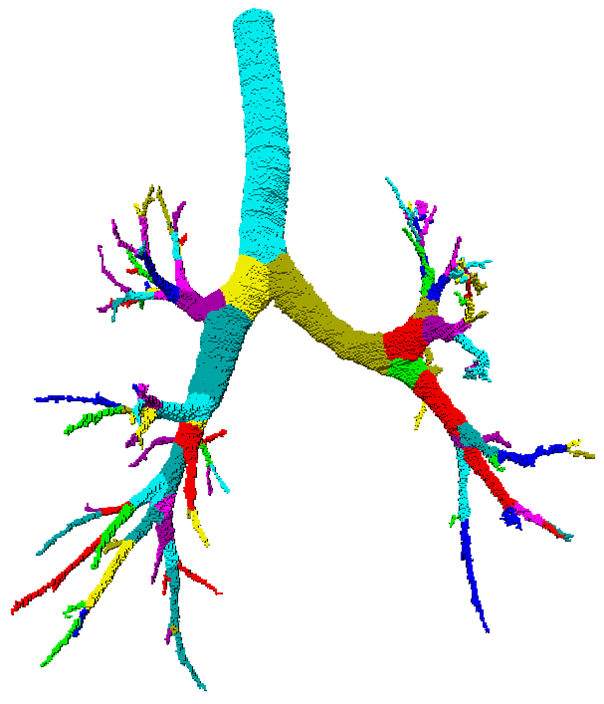
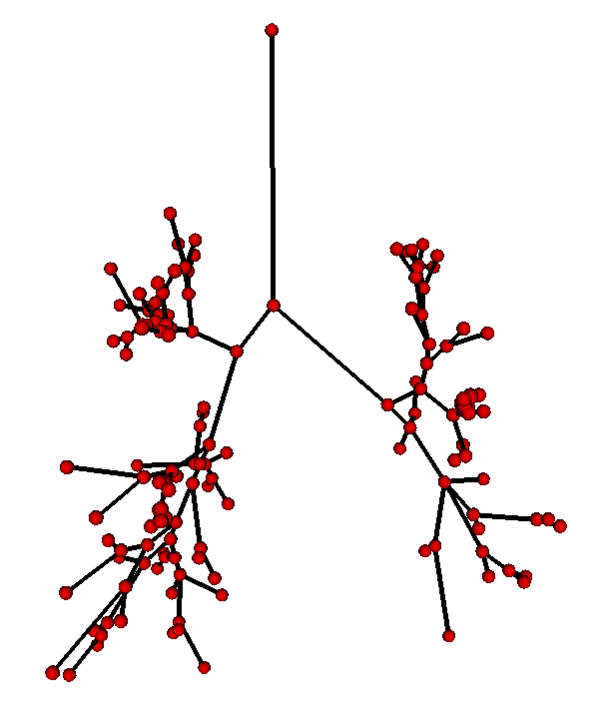
Partitioning centerlines in a formal tree data structure (left), partitioning segmented tree via isotropic label propagation (middle), and quantitative analysis formal XML tree with associated measurements (right).
