
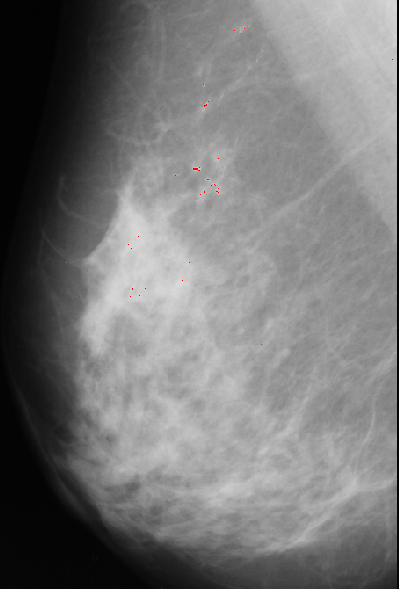
Figure 1.
Input:Given an image for example of industrial image of object with holes or such as a mammogram with the presence of microcalcifications of different size and shape (which can be introduced by simulation for the purpose of the project).
Method:Design a matched filter/Hough transform, capable of detecting them (in either space or Frequency space). The important feature is that the microcalcifications /circles are not of unique size or shape and are noisy. Possible aplication also of neural net. An alternative would be images of building with windows and counting windows.
Output:images with detection indicated
Q: What are breast microcalcifications?
A:Calcifications are small calcium deposits in the breast that can be detected only by mammography (breast x-ray). They are divided into two categories, macrocalcifications and microcalcifications.
Microcalcifications are tiny specks of calcium in the breast. These minute calcifications are usually found in an area of rapidly dividing cells. When many microcalcifications are seen in one area, they are referred to as a cluster and may indicate a small cancer. About half of the cancers detected by mammography appear as a cluster of microcalcifications.
Digital Mammography
Digital Mammography promises to be a significant improvement overconventional mammography in the early detection of breast cancer.
Digital mammography assists in the early detection of breast cancer
by incorporating modern electronics and
computers into x-ray mammography methods.
Instead of acquiring an image on film, it is collected electronically
and can be stored directly into a computer.
We have used images from http://peipa.essex.ac.uk/ipa/pix/mias/. The images were downloaded from the MIAS MiniMammographic Database, that were digitized at 200Ám (the original MIAS Database is digitised at 50Ám), with every image being 1024 pixels x 1024 pixels. Tipical mammograms used for image processing are digitized at 35Ám-70Ám and 4096 gray levels (12 bits), while we worked with mammograms digitized at 200Ám with 256 gray levels (8 bits) because of disk space. The use of these low-resolution images caused the disappearance of some microcalcifications that were present in the original images (digitized at 50Ám).
We have used several approaches to solve the problem. These algorithms are outlined bellow:
1.Unsharp Filtering
We used a variation of the unsharp masking method for enhancement of the microcalcifications in the original mammogram. The theory for unsharp filtering is that a low-pass filtered version of an image is subtracted from the image, leaving high frequency information. In this case, due to the small size of the microcalcifications (usually less than 5 pixels big), we achieved better results using a 3 x 3 mask for the low-pass filtering, followed by the image subtraction, thus, enhancing the high-frequency components of the image.
The unsharp filtering only enhances the high-frequency components of the image, thus, has to be followed by a thresholding that will segment the microcalcifications from the breast tissue. This thresholding was done manually and the values for the threshold variable ranged from 170 to 215 in the 4 images.
A result using this approach is presented bellow:


2.Edge detection
First we have applied an edge detector filter to the image.
| 3 | 3 | 3 |
| -5 | 0 | 3 |
| -5 | -5 | 3 |

There are many black pixels, and few bright pixels. The selected threshold is at the first position in the histogram where the difference between two consecutive positions is smaller then a given value. After that we eliminate the shapes with areas bigger than given threshold. The result of this approach is presented bellow:
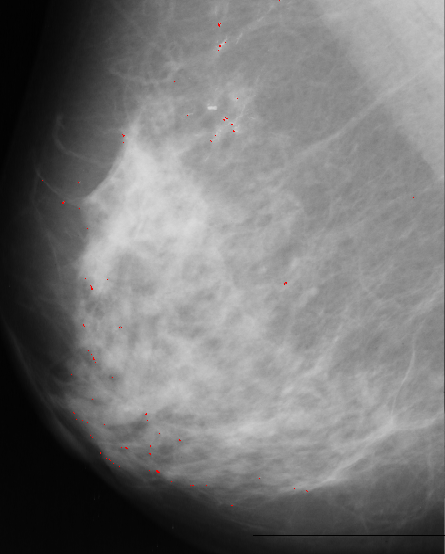
As we can see this approach is very noisy, contain many false-positives.
3.Median filter
Median filter approach is based on assumption that microcalcification are small spots with higher intensity then surrounding background. First we apply median filter to bright area of the original image. Median filter eliminates small isolated spots including microcalcifications. Next step is to bring them back by substracting filtered image from original one. Since we are only interested in spots brighter than background we keep only positive resulting values and set other to zero. Our next step is to match resulting image with 5x5 template. It has bell shape with value of 1 at the central point, which we expect to have a best fit at the position of microcalcification.
Template, multiplied with pixel value at current position, is substracted from median filtered image. The results are normalized, thresholded and added to
original image to point to possible position of microcalcification.
A result of this approach is:

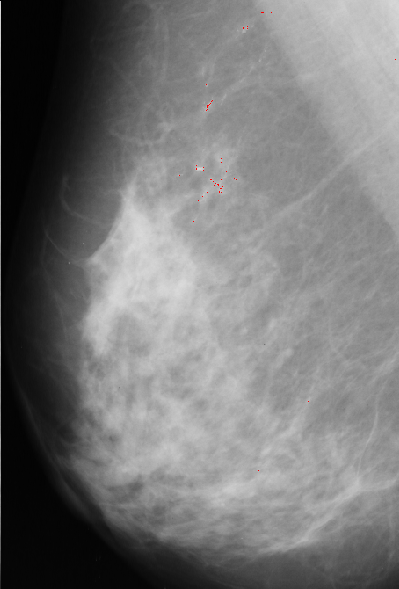
Our algorithms, due to the poor contrast of some areas of the images and the low-resolution of them, produced some false-positive results. The next step in our algorithms would be the use of some local information and object shape for the purpose of classifying the segmented objects in microcalcifications (benings and malignants) and artifacts, reducing the number of false-positives. Some of the possible features to be used for this purpose area :
The methods that we developed seem to detect the microcalcifications that were pointed by the MIAS database diagnosis (We used their results as ground truth for our problem), but also produced some false-positive. We believe that some of these false-positives were generated due to the low-resolution of the images. Further work would be necessary to get a better detection algorithm, maybe through some shape analysis or some local information (area surrounding the object).